Main Body
Prokaryotic gene regulation- Lac Operon
Prokaryotic gene regulation- Lac Operon
Dr.V.Malathi
Lactose Operon: An Inducible Operon
Francois Jacob & Jacques Monod. studied the mechanisms of transcriptional regulation in E. coli.
The lac operon consists of two main components namely the regulatory genes and structural genes .
The structural genes
The structural genes are lax Z, lac Y and lac A. These encode the three enzymes involved in lactose metabolism ; These are lac Z encodes an enzyme calledβ-galactosidase, which digests lactose into its two constituent sugars namely glucose and galactose. lac Y encodes the enzyme called permease that helps to transfer lactose into the cell and lac A encodes trans-acetylase; the relevance of which in lactose metabolism is not clearly understood.
The regulatory genes
In addition to the three protein-coding genes, the lac operon contains short DNA sequences that do not encode proteins, These sequences are called P (promoter), O (operator), and CBS (CAP-binding site) . These are binding sites for proteins involved in transcriptional regulation of the operon.
These sequences are called cis-elements as they are located on the DNA of the genes they regulate. On the other hand, the proteins that bind to these cis-elements are called trans-regulators . These are diffusible molecules and are not necessarily need to be encoded on the same piece of DNA as the genes they regulate.
One of the major trans-regulators of the lac operon is encoded by lacI. which codes for the lac I protein. Four identical molecules of lacI proteins assemble together to form a homotetramer called a repressor.
Negative regulation of the Lac Operon
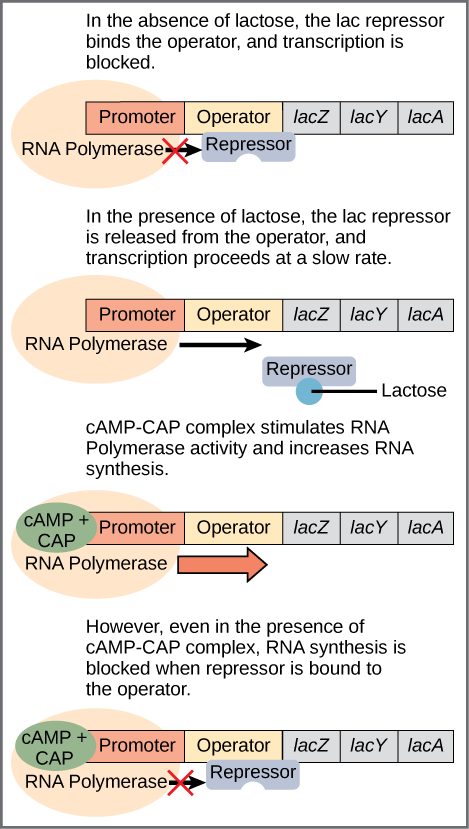
In the absence of Lactose and when glucose is available in the medium , the repressor binds to two operator sequences adjacent to the promoter of the lac operon. Binding of the repressor prevents RNA polymerase from binding to the promoter . Therefore, the operon will not be transcribed when the operator is occupied by a repressor. This is called negative regulation.
Positive Regulation
When the level of glucose in the medium is very low or non-existent and when lactose is available in the medium, the lac operon is induced to express the genes i.e., Only when glucose is absent and lactose is present will the lac operon be transcribed .When lactose is present, its metabolite, allolactose, binds to the lac repressor and changes its shape so that it cannot bind to the lac operator to prevent transcription. This is referred to as positive regulation
It should be mentioned that the lac operon is transcribed at a very low rate even when glucose is present and lactose absent.
Positive Regulation by CAP
CAP is a trans-factor called cAMP binding protein. CAP protein binds to a cis element within the lac promoter called CAP binding sequence (CBS). CBS is located very close to the promoter (P). CAP can bind to CBS only after it is binds with cAMP .When CAP is bound to at CBS, RNA polymerase is better able to bind to the promoter and initiate transcription. Thus, the presence of cAMP ultimately leads to a further increase in lac operon transcription.
The concentration of cAMP is inversely proportional to the abundance of glucose: when glucose concentrations are low, the enzyme called adenylate cyclase produces cAMP from ATP.E. coli prefers glucose over lactose, and so expresses the lac operon at high levels only when glucose is absent and lactose is present. The lac operon expressed at its highest levels only in the presence of lactose, and in the absence of glucose .
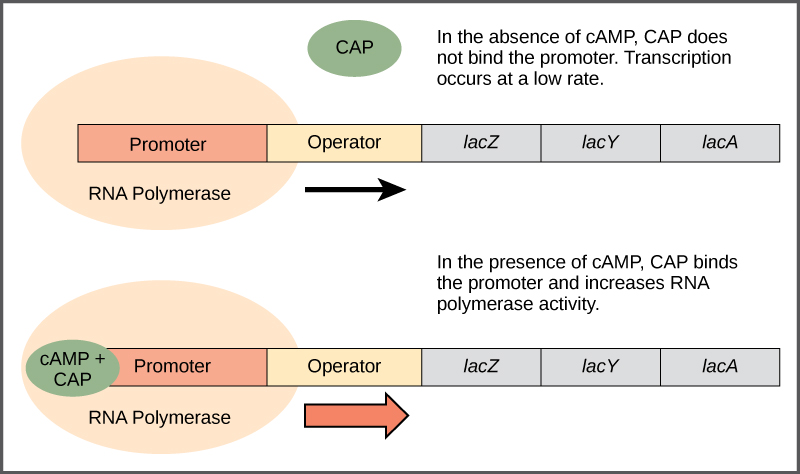
“cAMP binds to the CAP protein, a positive regulator” from Biology, an OpenStax resource is licensed under CC BY-SA 4.0
Here is a simulation on Gene machine-The lac operon. After playing with this simulation you will be able to
- Predict the effects on lactose metabolism when the various genes and DNA control elements are mutated (added or removed).
- Predict the effects on lactose metabolism when the concentration of lactose is changed.
- Explain the roles of LacI, LacZ, and LacY in lactose regulation.
Below is another simulation on Gene Expression Essentials. After interacting with the simulation you will be able to
- Explain the main sequence of events that occur within a cell that leads to protein synthesis.
- Predict how changing the concentrations and interactions of biomolecules affects protein production.
- Explain how protein production in a single cell relates to the quantity produced by a collection of cells.
Click on each link – for example when you click on Expression, that will be highlighted, click again on it to go further, you can use Home button to return.
“Gene Expression Essentials “ by PHET is licensed under CC BY 4.0

