Main Body
Prokaryotic Translation Elongation & Termination
Prokaryotic Translation Elongation & Termination
Dr.V.Malathi
Elongation
- Elongation includes repetitive cycles of decoding the m RNA , peptide bond formation, and translocation.
- Elongation begins as soon as the second codon of the ORF becomes accessible for reading by the next amino acylated tRNAs
- Elongation is facilitated by translation factors EF-Tu, EF-G ,EF-Ts EF-P and EF-4.
| Bacterial Elongation Factors | Function |
| EF-TU | mediates the entry of the aminoacyl t RNA into a free site of the ribosome |
| EF-TS | serves as the guanine nucleotide exchange factor for EF-Tu and catalyzes the release of GDP from EF-Tu. |
| EF-G | catalyzes the translocation of the tRNA and mRNA down the ribosome. |
| EF-P | possibly stimulates formation of peptide bonds and resolves stalls. |
| EF-4 | Proofreading |
- The second codon in the P site is recognized by aminoacylated -tRNAs. A ternary complex of aa-tRANA ,EF-Tu and GTP is attached to the P site .This interaction triggers GTP hydrolysis by EF-Tu. After Pi release, EF-Tu rearranges into the guanosine diphosphate (GDP)-bound form and releases the aa-tRNA.
- The amino group of the amino acid attached to the A-site tRNA makes a peptide bond with the carboxyl group of the amino acid attached to the P-site tRNA. This reaction is catalyzed by peptidyl transferase, an RNA-based ribozyme that is integrated into the 50S ribosomal subunit. Now the A site carries a peptide comprising of two amino acids and leaves the uncharged t RNA in the p site .
- During elongation the ribosomes move one codon along the mRNA in a process called Translocation. Translocation is promoted by EF-G at the cost of GTP hydrolysis .During each translocation event, a new charged tRNAs enter at the A site, then shift to the P site, and then finally to the E site for removal. Ribosomal movements are induced by conformational changes that advance the ribosome by three bases in the 3′ direction.
- This process of elongation continues until the ribosome arrives at the stop codon.
- Amazingly, the E. coli translation apparatus takes only 0.05 seconds to add each amino acid, meaning that a 200 amino-acid protein can be translated in just 10 seconds.
Termination
- Termination occurs when the ribosome encounters a stop codon UAG/UAA and UGA/UAA, respectively in the mRNA.
- These stop codons are recognized by the termination (or release) factors RF1 and RF2.
- Another termination factor, RF3, facilitates turnover of RF1 and RF2 but is not required for peptidyl-tRNA hydrolysis.
- The mechanism of termination encompasses three steps:
- recognition of the stop codon,
- hydrolysis of the ester bond of the peptidyl-tRNA (these two steps are accomplished by RF1 or RF2) and
- dissociation of RF1/RF2 with the help of RF3.
- The releasing factors instruct peptidyl transferase to add a water molecule to the carboxyl end of the P-site amino acid. This reaction forces the P-site amino acid to detach from its tRNA, and releases the newly made protein.
- The small and large ribosomal subunits dissociate from the mRNA and from each other; they are recruited almost immediately into another translation initiation complex. After many ribosomes have completed translation, the mRNA is degraded so the nucleotides can be reused in another transcription reaction.
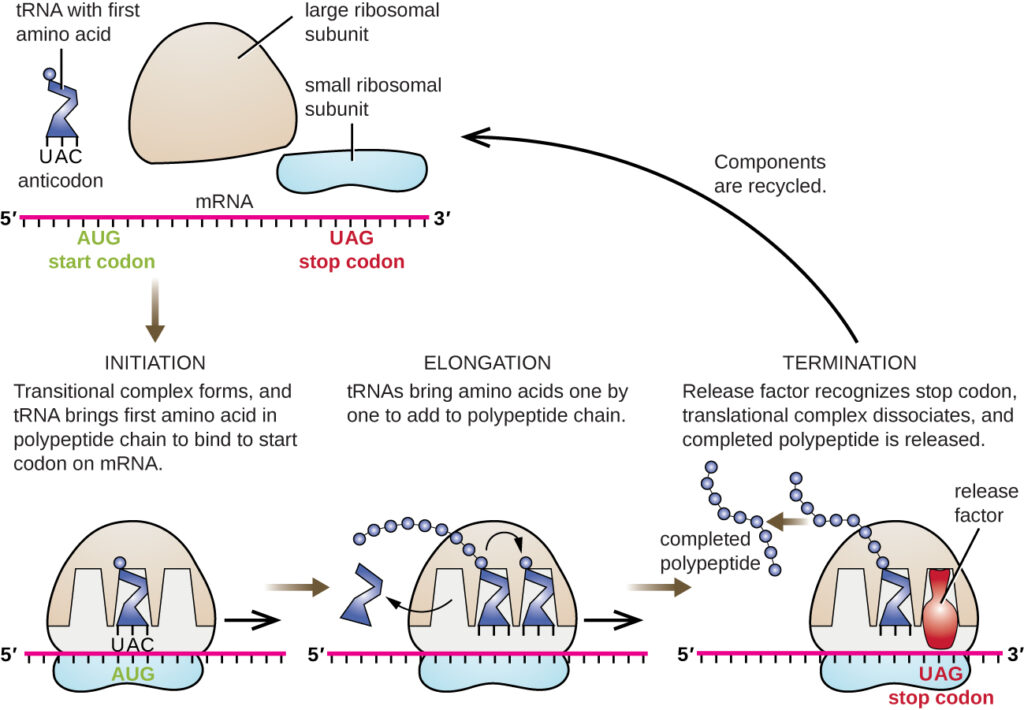
“Translation in bacteria” by Biology 2e. Provided by: OpenStax is licensed under CC BY 4.0

