Main Body
Prokaryotic gene regulation- Trp Operon
Prokaryotic gene regulation- Trp Operon
Dr.V.Malathi
The trp Operon: A Repressor Operon
Bacteria such as E. coli need amino acids like tryptophan to survive, which they can ingest from the environment.
E. coli can also synthesize tryptophan using enzymes that are encoded by five genes in the tryptophan (trp) operon .
If tryptophan is available in the environment, then there is no necessity for E. coli does to synthesize it and the trp operon is switched off. However, when tryptophan availability is low, the operon is turned on, transcription is initiated, the genes are expressed, and tryptophan is synthesized.
Like lac operon , trp operon also consists of structural genes and regulatory genes
The regulatory genes include
P/O genes : Promoter sequence and the operator sequence which is found in the promoter region.
trp L : Leader sequence : attenuator (A) sequence is found in the leader
The structural genes are
| trp Operon Gene | Gene Function |
| trp E | Gene for anthranilate synthetase subunit |
| trp D | Gene for anthranilate synthetase subunit |
| trp C | Gene for glycerolphosphate synthetase |
| trp B | Gene for tryptophan synthetase subunit |
| trp A | Gene for tryptophan synthetase subunit |
The trp operon is a repressible systems because the binding of the effector molecule to the repressor greatly increases the affinity of repressor for the operator . The repressor binds to the operator and stops transcription. Thus if tryptophan (effector) is available in the medium the repressors binds at the operator and repress the trp operon.
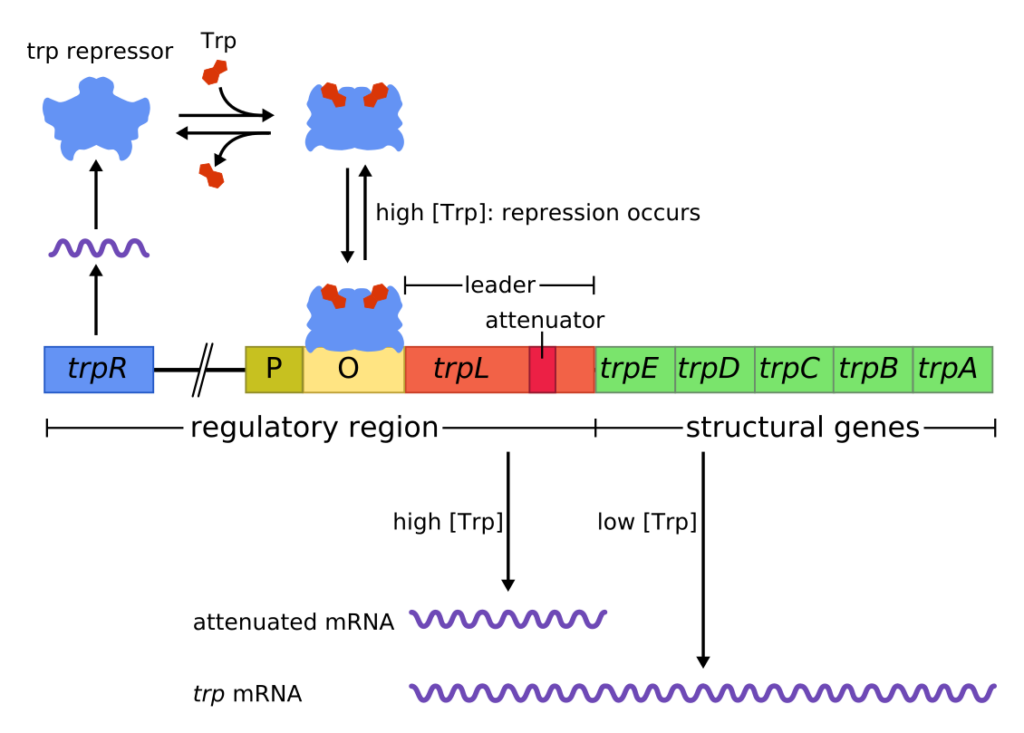
Histidine-Trpoperon by Wikimedia Commons licensed under CC BY-SA 3.0
Attenuation of the trp Operon
- The leader sequence (L) of the trp Operon lies before the trpE gene at its 5′ end . This sequence about 160 bp is size and controls the expression of the operon through a process called attentuation.
- This sequence has four domains (1-4). Domain 3 (nucleotides 108-121) of the mRNA can base pair with either domain 2 (nucleotides 74-94) or domain 4 (nucleotides 126-134).
- At high levels of tryptophan in the medium : domain 3 pairs with domain 4, a stem and loop structure forms on the mRNA and transcription stops.
- At low levels of tryptophan in the medium : domain 3 pairs with domain 2, then the stem and loop structure does not form and transcription continues through the operon, and all of the enzymes required for tryptophan biosynthesis are produced.
- Attenuators: Domain 4 is called the attenuator because its presence is required to reduce (attenuate) mRNA transcription in the presence of high levels of tryptophan.
- Domain 1 is also an important component of the attenuation process. The section of the leader sequence encodes a 14 amino acid peptide that has two tryptophan residues.
trp Operon Transcription Under High Levels of Tryptophan
- When the cellular levels of tryptophan are high, the levels of the tryptophan tRNA are also high.
- Immediately after transcription, the mRNA moves quickly through the ribosome complex translating the leader peptide .
- Translation is quick because of the high levels of tryptophan tRNA.
- Domain 2 becomes associated with the ribosome complex.
- As result the domain 3 binds with domain 4, and
- transcription is attenuated because of the stem and loop formation.
trp Operon Transcription Under Low Levels of Tryptophan
- Under low cellular levels of tryptophan, the translation of the peptide on domain 1 is slow.
- Domain 2 does not become associated with the ribosome.
- Rather domain 2 associates with domain 3.
- This structure permits the continued transcription of the operon.
- Then the trpE-A genes are translated, and the biosynthesis of tryptophan occurs.
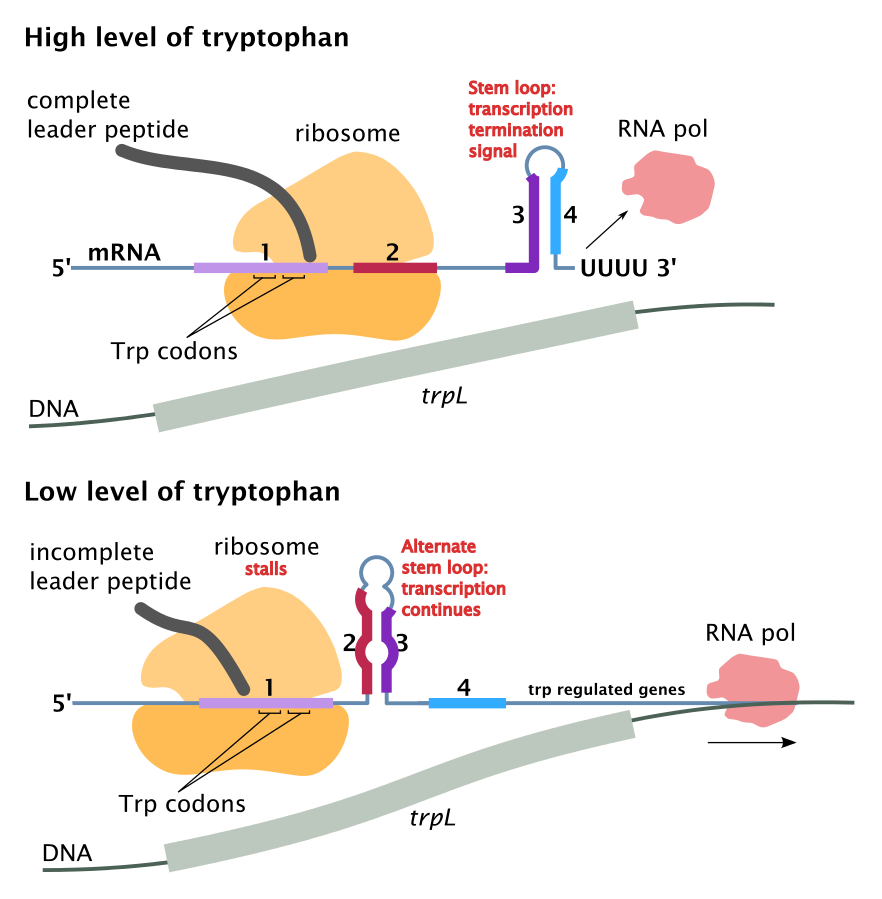
Trp operon attenuation by Wikimedia commons is licensed under CC BY-SA 3.0

