Main Body
Eukaryotic Transcription gene regulation
Eukaryotic Transcription gene regulation
Dr.V.Malathi
Transcriptional Regulation in Eukaryotes
There are three classes of control elements in eukaryotes:
cis-acting DNA element
cis-acting DNA elements- enhancers and silencers
•An enhancer DNA sequence (or positive regulatory element) turns a gene ON.
•When the activator is bound to the enhancer, RNA polymerase is more highly attracted to the gene.
•enhancers are located upstream or downstream of the promoter region,
•Multiple regulatory proteins bound to binding sites in an enhancer can form a large, complex enhancesosome that has varying affinity for RNA polymerase, depending on its size and exact composition.
•The enhanceosome can both recruit additional co-activators and facilitate chromatin remodeling.
•A silencer DNA sequence (or negative regulatory element) turns a gene OFF or reduces its rate of transcription.
•When the repressor is bound to the silencer, RNA polymerase cannot attach and transcribe the gene.
•silencers are located downstream of a promoter.
Trans-acting DNA element
Trans-acting factors:
Post-Translational Control of Gene Expression
- RNA is transcribed, but must be processed into a mature form before translation can begin. This processing is called post-transcriptional modification.
- This post-transcriptional step can also be regulated to control gene expression in the cell.
- In eukaryotic RNA transcript often contains regions, called introns, that are removed prior to translation.
- The regions of RNA that code for protein are called exons .
- By a process called splicing , the RNA is processed and the introns are removed and exons are ligated together.
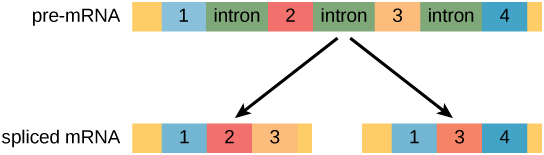
“Pre-mRNA can be alternatively spliced to create different proteins” by Shelli Carter and Lumen Learning. is licensed under CC BY 4.0
ALTERNATIVE RNA SPLICING
- Alternative RNA splicing is a mechanism that allows different protein products to be produced from one gene when different combinations of introns, and sometimes exons, are removed from the transcript.
- Alternative splicing acts as a mechanism of gene regulation,
- The frequency of different splicing alternatives controlled by the cell as a way to control the production of different protein products in different cells or at different stages of development.
- 70 percent of genes in humans are expressed as multiple proteins through alternative splicing.
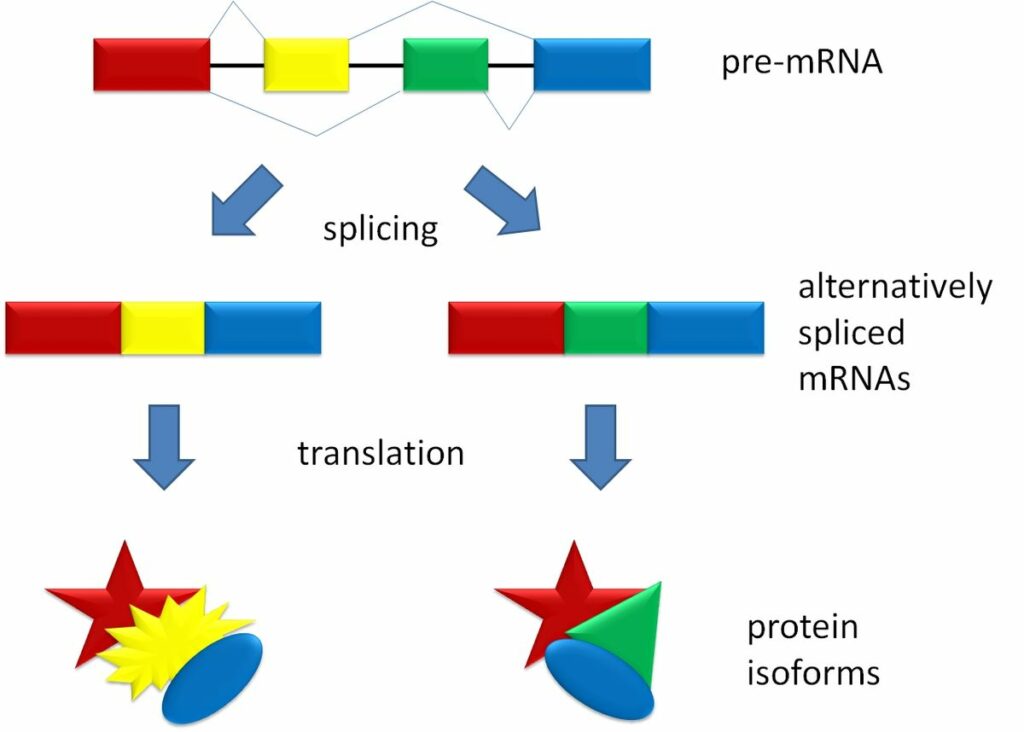
“Splicing_overview.jpg” by Agathman is licensed under CC BY-SA 3.0
Control of RNA Stability
- The 3′ and 5′ ends of the mRNA is protected before it leaves the nucleus,
- .The 5′ end of the mRNA is protected by a 5′ cap, which is usually composed of a methylated guanosine triphosphate molecule (GTP).
- The poly-A tail protects the 3′ end. It is usually composed of a series of adenine nucleotides.
- The length of time that the RNA resides in the cytoplasm can be controlled.
- Each RNA molecule has a defined lifespan and decays at a specific rate. This rate of decay determine how much protein is in the cell. For example , If the decay rate is increased, the RNA will not exist in the cytoplasm as long, shortening the time for translation to occur.
- This rate of decay is referred to as the RNA stability
- Proteins, called RNA-binding proteins, or RBPs, can bind to the regions of the RNA just upstream or downstream of the protein-coding region. These regions in the RNA that are not translated into protein are called the untranslated regions, or UTRs.
- These regions regulate mRNA localization, stability, and protein translation. The region just before the protein-coding region is called the 5′ UTR, whereas the region after the coding region is called the 3′ UTR .
- The binding of RBPs to these regions can increase or decrease the stability of an RNA molecule, depending on the specific RBP that binds.
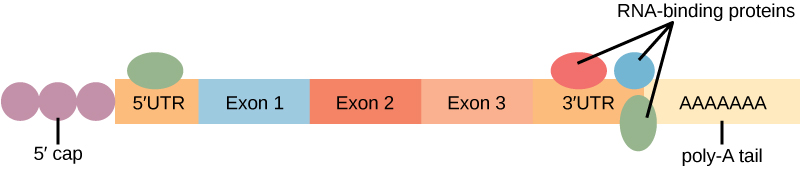
“The protein-coding region of mRNA is flanked by 5′ and 3′ untranslated regions (UTRs). The presence of RNA-binding proteins at the 5′ or 3′ UTR influences the stability of the RNA molecule.” by Shelli Carter and Lumen Learning is licensed under CC BY 3.0
microRNAs
- microRNAs, or miRNAs, are short RNA molecules that are only 21–24 nucleotides in length.
- The miRNAs are made in the nucleus as longer pre-miRNAs.
- A protein called dicer chop these pre-miRNAs are chopped into mature miRNAs .
- Mature miRNAs recognize a specific sequence and bind to the RNA and associate with a ribonucleoprotein complex called the RNA-induced silencing complex (RISC).
- RISC binds along with the miRNA to degrade the target mRNA. Together, miRNAs and the RISC complex rapidly destroy the RNA molecule.
The processing of the RNA involving the removal of introns and ligation of exons
The regions of m RNA that are not translated in to protein.

