7 Biosystematics
7.3 Phylogenetics
Phylogenetics
Dr V Malathi
The study of evolutionary relationships among organisms is called Phylogenetics. It reconstructs the evolutionary history of species and charts their relationships in a phylogenetic tree using genetic, morphological, and biochemical data. Understanding the relationships between species, the history of their evolutionary divergence, and the evolution of features across time is made easier by phylogenetics.
Phylogenetic Trees
To illustrate the evolutionary relationships and paths between organisms, scientists employ a particular kind of model known as a phylogenetic tree also called as evolutionary trees. It is an illustration of the evolutionary links between individuals or groups of organisms . As the suggested linkages cannot be verified in the past, scientists view phylogenetic trees as a hypothesis of the evolutionary past.
Terminology that characterizes a phylogenetic tree.
A common ancestor is represented by a single lineage at the base of many phylogenetic trees. Such trees are referred as rooted phylogenetic tree, indicating that all of the creatures shown in the diagram are related to a single ancestral lineage, usually drawn from the bottom or left. The three domains of bacteria, archaea, and eukarya branch off from a single point in the rooted phylogenetic tree.In this image, the little branch that plants and animals—including humans—occupy demonstrates how recent and insignificant these groupings are in relation to other species. Unrooted phylogenetic trees reveal links between species but not a common ancestor.

Clade or monophyletic group
The branch path depicting an ancestor–descendant relationship on a tree is called a lineage.

Tree topology
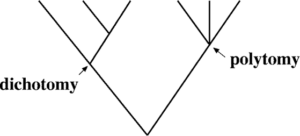
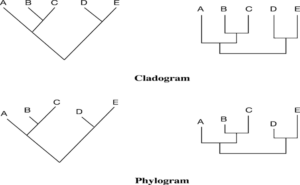
Forms of Tree Representation
Phylogram and Cladogram
Molecular Phylogenetics:
enables researchers to track lineages back to common ancestors by analyzing evolutionary relationships using DNA, RNA, or protein sequences.
Molecular techniques have transformed phylogenetics by offering information that helps elucidate connections in situations when morphological evidence is unclear or lacking.
Methods of Building Phylogenetic Trees
- Distance Methods:
- These methods create a tree based on overall similarity.These techniques determine the genetic distance between two sequence pairs.
- Neighbor-joining is a popular distance method that reduces the tree’s overall branch length.
- Maximum Parsimony:
- This approach assumes that simpler evolutionary pathways are more plausible than complex ones, and it looks for the tree with the fewest evolutionary changes.
- Maximum Likelihood and Bayesian Methods:
- The Bayesian and maximum likelihood statistical techniques determine the likelihood of a specific tree given an evolutionary change model.
While Bayesian approaches predict a range of plausible topologies, Maximum Likelihood uses sequence data to determine which tree has the highest likelihood.
- The Bayesian and maximum likelihood statistical techniques determine the likelihood of a specific tree given an evolutionary change model.
Applications of Phylogenetics
- Classification and Taxonomy: Contributes to the improvement of the biological categorization system by grouping species according to evolutionary links.
- Understanding Evolutionary History: The chronology and order of evolutionary events, including speciation and trait development, can be known by an understanding of evolutionary history.
- Tracking Disease Evolution: This method is used in epidemiology to investigate the causes and dissemination of infections, including monitoring the genetic development of viruses such as SARS-CoV-2 and HIV.
- Conservation Biology: Assists in identifying evolutionary distinct species that, because of their distinct genetic heritage, may be conservation priorities.
- Comparative Genomics: Phylogenetics helps to compare genomes across species, identify conserved genes, and to study gene function and evolution.
Test your Understanding

