5 Genetics and Evolution
5.5 Multiple allelism, Recombination, Sex linkage
Mutliple alleles, Recombination, Linkage
Dr V Malathi
Multiple allelism
According to Mendel, only two alleles, one dominant and one recessive, could exist for a given gene.
Most diploid organisms including humans can only have two alleles for a given gene
However multiple alleles may exist at the population level .
In such cases many combinations of two alleles are observed.
When many alleles exist for the same gene, it is convention to denote the most common phenotype or genotype as the wild type (often abbreviated “+”);
All other phenotypes or genotypes are considered variants of this standard, meaning that they deviate from the wild type. The variant may be recessive or dominant to the wild-type allele.
Example : Coat color in rabbits .
Here, four alleles exist for the c gene.
- The wild-type version, has Genotype : C+C+, is expressed as brown fur.
- The chinchilla , genotype : has cchcch, is expressed as black-tipped white fur.
- The Himalayan phenotype, has genotype : chch, has black fur on the extremities and white fur elsewhere.
- The albino, or “colorless” phenotype, genotype cc, is expressed as white fur.
In cases of multiple alleles, dominance hierarchies can exist.
In above case, the wild-type allele is dominant over all the others, chinchilla is incompletely dominant over Himalayan and albino, and Himalayan is dominant over albino.

“Four different alleles of rabbit” by Lumen learning is licensed under CC BY 4.0
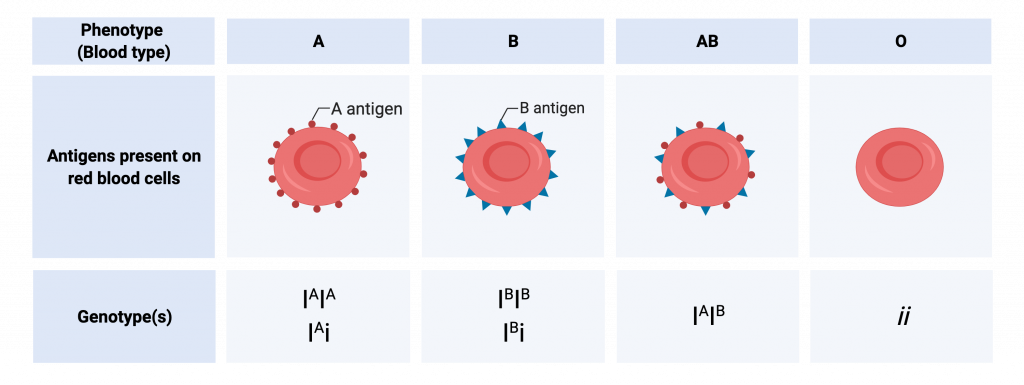
Another example for multiple allelism is ABO blood grouping in humans
The ABO blood groups is determined according to the type of the antigen protein expressed on the surface of the Red Blood Cells .The three alleles: IA, IB, and i determine the blood group
- The A blood group blood RBCs express the Antigen A on their surface ; Expressed by the alleles IA IA and IA i
- The B blood group blood RBCs express the Antigen B on their surface ; Expressed by the alleles IB IB and IB i
- The A Bblood group blood RBCs express both the Antigen A and B on their surface ; Expressed by the alleles IA IB
- The O blood group blood RBCs do not express any Antigen A on their surface ; These possess the alleles ii
Alleles IA and IB are codominant with respect to one another, and both are dominant to i. These three alleles in the population result in six genotypes, and four phenotypes.
” ABO blood system in humans “ by Melissa Hardy is licensed under CC BY-NC 4.0
Watch the video from Amoeba Sisters on Multiple alleles
Genetic Recombination
Genetic recombination involves the exhange of genetic material between organisms. This leads to the production of offspring with combinations of traits that differ from either parent.
In eukaryotes ,genetic recombination occurs during meiosis .
Recombination can be classified in to two types
- Interchromosomal recombination : This occurs through independent assortment of alleles, which are on different but on homologous chromosomes.
- Intrachromosomal recombination : Occurs through crossing over
Crossing over
Each parent cell has pairs of homologous chromosomes,
• One homolog from the Paternal and one from the maternal origin.
• These chromosomes are replicated before cell division
• In prophase I of meiosis, the replicated homologous pair of chromosomes comes together and this process is called Synapsis.
• Sections of the paired chromosomes are exchanged.
• This exchange occurs by a process called crossing over.
• After crossing over, the resultant chromosomes are neither resemble entirely maternal nor entirely paternal, but contain genes from both parents.
• This ensures genetic variation in sexually reproducing organisms
The main features of crossing over are given below:
1.Crossing over takes place during meiotic prophase, i.e., during pachytene. Each pair of chromosome has four chromatids at that time.(Tetrad)
2. Crossing over occurs between non-sister chromatids.
3. One chromatid from each of the two homologus chromosomes is involved in crossing over.
4. Each crossing over involves only two of the four chromatids of two homologus chromosomes.
5.Rarely double or multiple crossing over may involve all four, three or two of the four chromatids.
6. Crossing over leads to re-combinations or new combinations.
7.Crossing over generally yields two recombinant types or crossover types and two parental types or non-crossover types.
6. Crossing over generally leads to exchange of equal segments or genes and recombination is always reciprocal. However, unequal crossing over has also been reported.
7. The value of crossover or recombinants may vary from 0-50%.
Chiasma and Crossing Over
• The point of exchange of segments between non-sister chromatids of homologous chromosomes during meiotic prophase is called chiasma (pleural chiasmata).
• It is the place where crossing over takes place.
• Depending on the position, chiasma is of two types, viz., terminal and interstitial.
• When the chiasma is located at the end of the pairing chromatids, it is known as terminal chiasma
• when it is located in the middle part of non-sister chromatids, it is referred to as interstitial chiasma.
• Later on interstitial chiasma is changed to terminal position by the process of chiasma terminalization.
• The number of chiasma per bivalent may vary from one to more than one depending upon the length of chromatids.
Chiasma Terminalization
• The movement of chiasma away from the centromere and towards the end of tetrads is called terminalization.
• The total number of chiasmata terminalized at any given stage or time is known as coefficient of terminalization.
• Generally, chiasma terminalization occurs between diplotene and metaphase I
Mechanism of Crossing Over
• Crossing over begins with a double strand break in one of the DNA molecules.
• This leads to two hanging single-stranded regions
• These single strand regions get coated with proteins called recombinase that catalyze recombination.
• The first invading strand behaves like a primer and this synthesizes a double stranded region for itself using one strand of its non-sister chromatid as a template.
• This leads to its complementary strand getting displaced and base pairing with the second single stranded region that was initially generated by the exonuclease.
• Ultimately, this results in two strands being exchanged with the formation of a cross-like structure called the Holliday junction

Repair of the gap can lead to crossover (CO) or non-crossover (NCO) of the flanking regions. CO recombination is thought to occur by the Double Holliday Junction (DHJ) model, illustrated on the right. NCO recombinants are thought to occur primarily by the Synthesis Dependent Strand Annealing (SDSA) model, illustrated on the left. Most recombination events appear to be the SDSA type.
“Homologous Recombination” by Harris Bernstein, Carol Bernstein and Richard E. Michod is licensed under CC BY 3.0
Linkage
There are more genes than the chromosome
• Therefore each chromosome contains more than one gene.
• The genes for different characters may be either situated in the same chromosome or in different chromosome
• When the genes are situated in different chromosomes the characters they control appear in the next generation either together or apart. i.e they assort independently according to Mendel’s Law of Independent Assortment.
- But if genes are situated in the same chromosome and if they are fairly close to each other ,they tend to be inherited together. This is called Linkage
• The linked genes do not assort independently but tend to stay together in the same combination as they were in the parents. - Genetic linkage describes the tendency of allele that are located close together on a chromosome to
be inherited together during the meiosis phase of sexual reproduction.
• Genes whose loci are nearer to each other are less likely to be separated onto different chromatids during chromosomal crossover.
• Such genes are said to be genetically linked i.e., the nearer two genes are on a chromosome, lesser is the chance of a crossing occurring between them, and therefore they are more likely they are to be
inherited together. - Genetic linkage was first discovered by the British geneticists William Bateson, Edith Rebecca Saunders and Reginald Punnett shortly after Mendel’s laws were rediscovered.
• The study about genetic linkage was expanded by the work of Thomas Hunt Morgan.
• Morgan observed that the amount of crossing over between linked genes differs
• This observation led to the idea that crossover frequency might indicate the distance separating genes on the chromosome.
Chromosomes Theory of Linkage
Morgan & castle formulated the chromosome theory of linkage which states that :-
1. The genes which show the phenomenon of linkage are situated in the same chromosomes and inherited together
during the process of inheritance.
2. The distance between the linked genes determines the strength of linkage. The closely located genes show strong linkage than the widely located genes which show the weak linkage.
3. The genes are arranged in linear fashion in the chromosomes.
Genetic maps or Linkage maps
Alfred Sturtevant (student of Morgan’s) first developed genetic maps.These are also known as linkage maps.
• He proposed that the greater the distance between linked genes, the greater the chance that non-sister
chromatids would cross over in the region between the genes.
• By calculating the number of recombinants it is possible to measure the distance between the genes.
• This distance is expressed in terms of a genetic map unit (m.u.), or a centimorgan
• It is defined as the distance between genes for which one product of meiosis in 100 is recombinant.
A recombinant frequency (RF) of 1% is equivalent to 1 m.u.
• This equivalence approximation holds good only for small percentages;
• The largest percentage of recombinants cannot exceed 50% ,which would be the situation where the two
genes are at the extreme opposite ends of the same chromosomes
Types of Linkage:
Linkage is of two types, complete and incomplete.
1. Complete Linkage (Morgan, 1919):
The genes located on the same chromosome do not separate and are inherited together over the generations due to the absence of crossing over.
Complete linkage allows the combination of parental traits to be inherited as such. It is rare but has been reported in male Drosophila and some other heterogametic organisms
Example:
• In Drosophila, genes of grey body and long wings are dominant over black body and vestigial (short)
wings.
• If pure breeding grey bodied long winged Drosophila (GL/ GL) flies are crossed with black bodied vestigial winged flies (gl/gl), the F2shows a 3 : 1 ratio of parental phenotypes (3 grey body long winged and one black body vestigial winged).
• This is explained by assuming that genes of body colour and wing length are found on the same chromosome and are completely linked.
Incomplete Linkage:
• Genes present in the same chromosome have a tendency to separate due to crossing over and hence produce recombinant progeny besides the parental type.
• The number of recombinant individuals is usually less than the number expected in independent assortment.
• In independent assortment all the four types (two parental types and two recombinant types) are each 25%.
• In case of linkage, each of the two parental types is more than 25% while each of the recombinant types is less than 25%
Example
A red eyed normal winged or wild type dominant homozygous female Drosophila is crossed to homozygous recessive purple eyed and vestigial winged male. The progeny or F1 individuals are heterozygous red eyed and normal winged. F1 female flies are test crossed with homozygous recessive males. It does not yield the ratio of 1: 1: 1: 1. Instead the ratio comes out to be 9: 1: 1: 8. This shows that the two genes did not segregate independently of each other.
Only 10.7% recombinant types were observed which is quite different from 50% recombinants in case of independent assortment. This shows that in the oocytes of the F1 , generation only some of the chromatids undergo crossover while the majority is preserved intact. This produces 90.3% parental types in the progeny
Test your Understanding

